Neuroscience
- Here's What We Get Completely Wrong When We're Judging The Difficulty Of Anagrams
We think pronounceable anagrams are easier, but they're harderWhen you're trying to solve an anagram (that is, re-arranging a jumble of letters to form a word), sometimes the string of letters looks like complete gobbledygook and impossible to...
- Three-person Groups Best For Problem-solving
Individuals may outperform groups when it comes to brainstorming for ideas (see earlier post), but for logic-based problem solving, it seems three-person groups work best. That’s according to Patrick Laughlin and colleagues who tested 760 students on...
- Monopoly Is A Perfect Example Of Embodied Cognition
I teach a Matlab programming class. The main project I get people to work on is programming up a version of the game Monopoly. It’s a great project, I think, because it makes you use all the things Matlab is good at (loops, matrices, data in and output,...
- Float Like A Butterfly; Sting Like A Bee.
Your hands can't hit what your eyes can't see. For this reason, we have to be careful when feeding numbers into our computer. Check out what happens when you ask a basic math question to Python: Pictured: the folly of man Huh, that's strange....
- De Brujin Graphs And Velvet Optimiser
I'm working with the Velvet Assembler as part of my virus identification project. When I'm not trying to write a perl module to complement the already-bulky script files that I'm working with, I like to do something that I usually wish...
Neuroscience
Longest Common Subsequence and NumPy
Perl may be crafty and efficient like a ninja, Ruby may be written like a prose or work of fiction, but, for most purposes, Python, with its simplicity and elegance, is usually my weapon of choice when it comes to programming languages. (To be frank, as long as it's not some cryptic code like Fortran that should probably be waiting for the rain to wash it away, it floats my boat.) With my knack for mathematics, I had been reconstructing various equations and theorems from scratch in most of my scripts. Recently, I've begun to embrace NumPy to give me more functionality for purposes like matrices and arrays, but also that I can do all the things my MATLAB friends do without too much effort to learn extra languages.
(In fact, while we're at it, let's just put everything in Python! Python-Excel, Python-sql, import everything!)
Back in my pancake post, I talked about how you can use a simple two-step algorithm for sorting out a string of numbers. In this post, however, I'm going to talk about sorting through two different string of letters to find their longest common subsequence. Unlike the challenge of the longest common substring, the subsequence need not consist solely of letters that are adjacent to one another, but can contain letters separated. So, this means that the longest common subsequence between "AACTTG" and "ACTGG" would be not be "ACT", but "ACTG".
I found this problem interesting because it gave me the chance to flex my NumPy muscles and look for more "indirect" ways of solving a problem rather than using a brute-force Ctrl+F-esque approach that wipes away your entire RAM when you search through strings longer than 30 characters.
Fret not! For we can construct a matrix of some sort to help us with this issue. In bioinformatics, we can solve this problem using a scoring matrix. By taking advantage of the set of possible DNA bases {A, C, T, G}:
I wrote a solution from this method (drawing heavily upon other sources) to solve this problem here, although I'm still fixing up some issues in it from converting between different syntaxes and formats.
(In fact, while we're at it, let's just put everything in Python! Python-Excel, Python-sql, import everything!)
Back in my pancake post, I talked about how you can use a simple two-step algorithm for sorting out a string of numbers. In this post, however, I'm going to talk about sorting through two different string of letters to find their longest common subsequence. Unlike the challenge of the longest common substring, the subsequence need not consist solely of letters that are adjacent to one another, but can contain letters separated. So, this means that the longest common subsequence between "AACTTG" and "ACTGG" would be not be "ACT", but "ACTG".
I found this problem interesting because it gave me the chance to flex my NumPy muscles and look for more "indirect" ways of solving a problem rather than using a brute-force Ctrl+F-esque approach that wipes away your entire RAM when you search through strings longer than 30 characters.
Fret not! For we can construct a matrix of some sort to help us with this issue. In bioinformatics, we can solve this problem using a scoring matrix. By taking advantage of the set of possible DNA bases {A, C, T, G}:
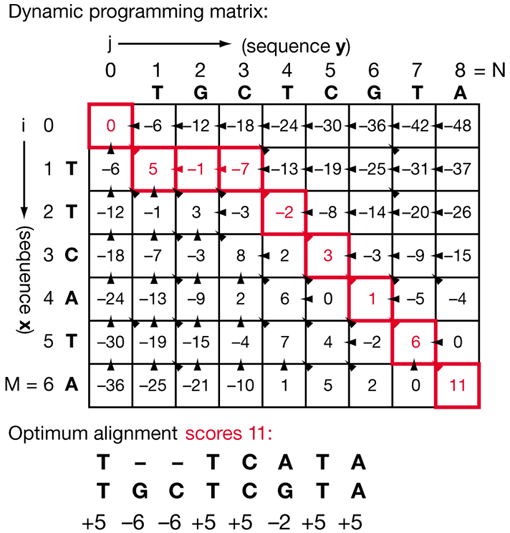 |
Simply place your two DNA strings on the axes and move each number in the grid from the top-left to bottom-right . If there is a match in the base between the two strings at a certain location, add one to the number. Then follow your path form highest to lowest value. (source) |
This is known as the Traceback approach, and we can optimize it further with hashes for lengths and in other ways.
I wrote a solution from this method (drawing heavily upon other sources) to solve this problem here, although I'm still fixing up some issues in it from converting between different syntaxes and formats.
- Here's What We Get Completely Wrong When We're Judging The Difficulty Of Anagrams
We think pronounceable anagrams are easier, but they're harderWhen you're trying to solve an anagram (that is, re-arranging a jumble of letters to form a word), sometimes the string of letters looks like complete gobbledygook and impossible to...
- Three-person Groups Best For Problem-solving
Individuals may outperform groups when it comes to brainstorming for ideas (see earlier post), but for logic-based problem solving, it seems three-person groups work best. That’s according to Patrick Laughlin and colleagues who tested 760 students on...
- Monopoly Is A Perfect Example Of Embodied Cognition
I teach a Matlab programming class. The main project I get people to work on is programming up a version of the game Monopoly. It’s a great project, I think, because it makes you use all the things Matlab is good at (loops, matrices, data in and output,...
- Float Like A Butterfly; Sting Like A Bee.
Your hands can't hit what your eyes can't see. For this reason, we have to be careful when feeding numbers into our computer. Check out what happens when you ask a basic math question to Python: Pictured: the folly of man Huh, that's strange....
- De Brujin Graphs And Velvet Optimiser
I'm working with the Velvet Assembler as part of my virus identification project. When I'm not trying to write a perl module to complement the already-bulky script files that I'm working with, I like to do something that I usually wish...
